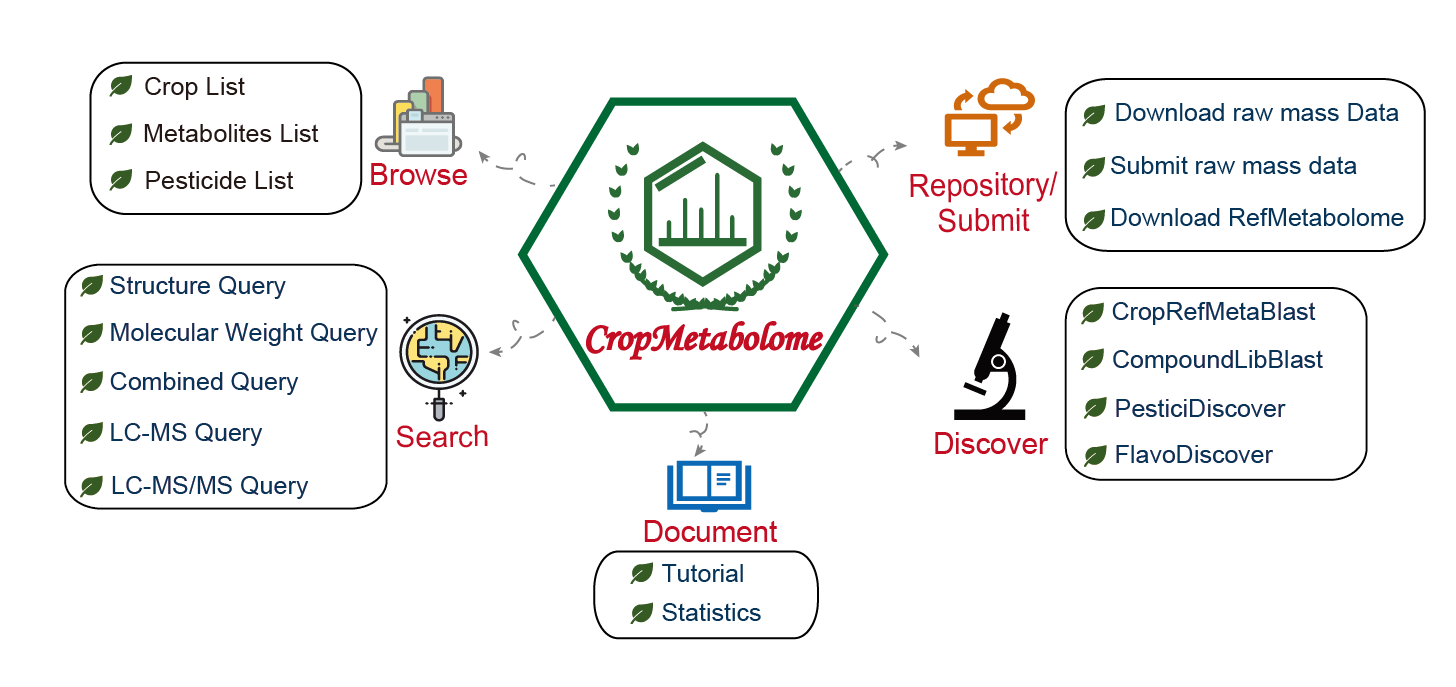
Overview
CropMetabolome is an integrated platform for crop metabolome databases and analysis tools that is dedicated to metabolic profiling, metabolite identification, referencing
standards, and repository and dissemination of crop metabolomics data. It currently contains three functional components:
1) metabolite reference libraries (MS/MS spectral tag [MS2T] libraries) for major crops (currently fifty-nine available) and the associated tool 'CropMetaDiscover' to annotate crop samples;
2) a repository for dissemination of curated crop metabolomics data;
3) analytical tools 'LC-MS Query', 'CropMetaDiscover', 'CompoundLibBlast', 'FlavoDiscover' and 'PesticiDiscover', for crops metabolite profiling and discovery, and pesticidal residue detection for toxicity evaluation.
Quick Search
Search by CropMetabolome ID, compound name, SMILES, molecular formula, and InChI.
You can input key fields of data entries including ID (Metabolite ID or compound ID), Name, Formula, InChI or SMILES in the search bar for quick search. The sub-query box will provide some options to search in metabolites, pesticides, compounds, and spectrum libraries in CropMetabolome.

Entrance for Quick Search
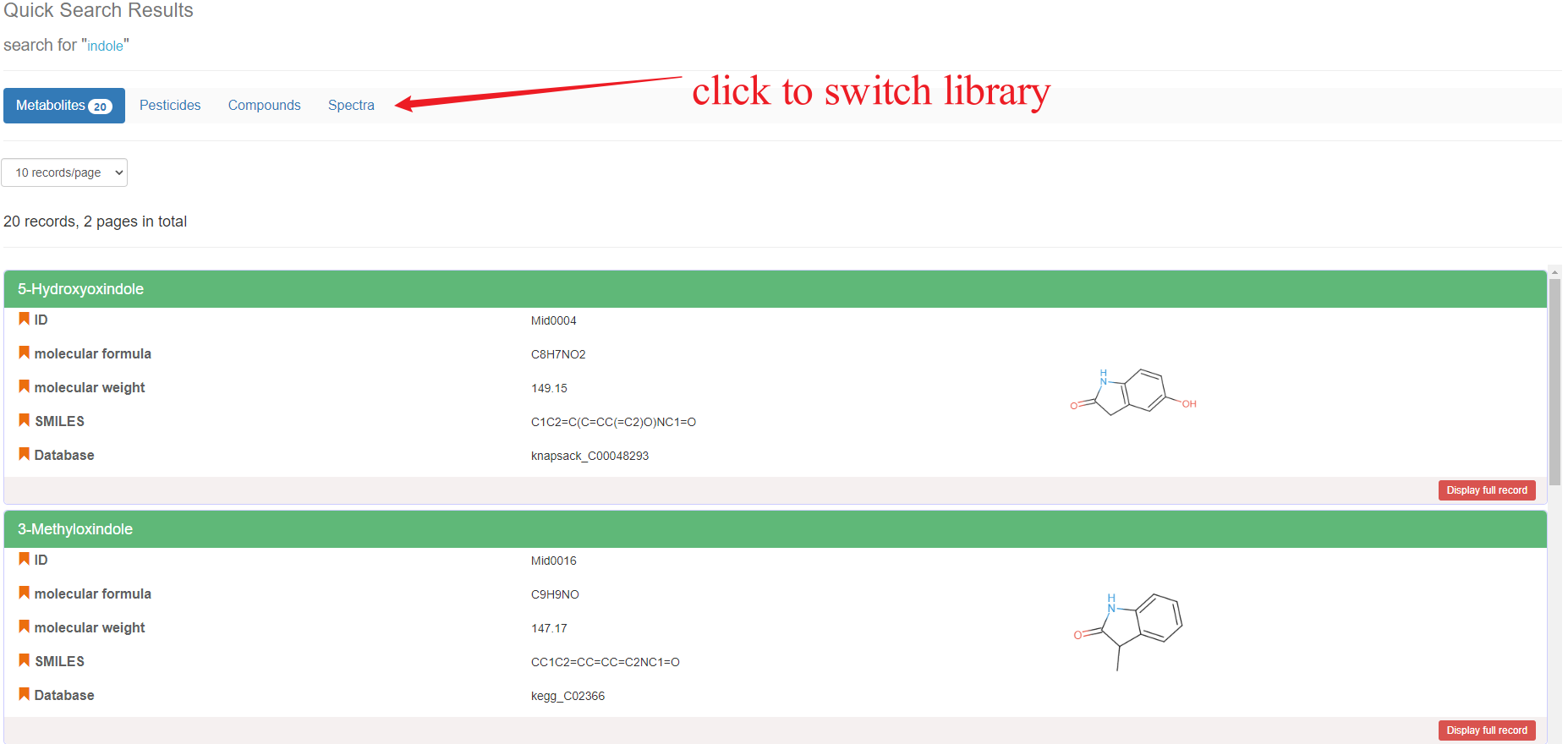
Quick Search Result
Browse
The ‘Browse’ module is designed for users to browse crop metabolites and compound data. Users can browse the webservice via clicking the "Browse" button on the homepage, and then access specific data via multiple options. Each data entry assigned unique CropMetabolome identifiers has a dedicated summary information card and detailed information page. And users can access entries of their interest quickly by a sub-query box on the browsing page.
1 Browse crop list
Usage
Users can click on the "Crop List" bar to access the Crop List webpage. The Crop List webpage displays information including the “Image,” “Common Name,” “Scientific Name,” “Metabolite Info,” and “Ref Metabolome” of the crops available in our database.
By clicking the "Display full record" button in the “Metabolite Info” column of the Crop List, users can access a new webpage to see all the metabolites that have been identified for the corresponding species. Detailed information of each metabolite can be accessed by clicking the "Display full record" button beneath each metabolite on the new webpage.
By clicking the records in the “Ref Metabolome” column of the Crop List webpage, users can download the reference metabolome in the .msr format for each crop.
Entrance for browsing crop list
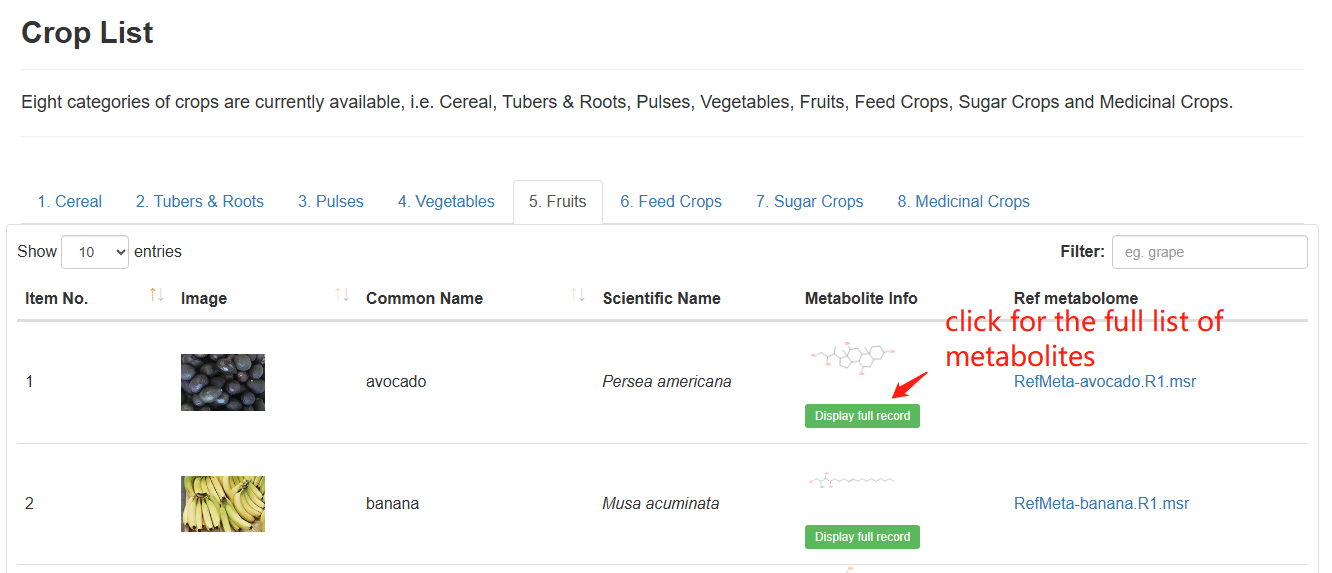
Overview of the CropList.
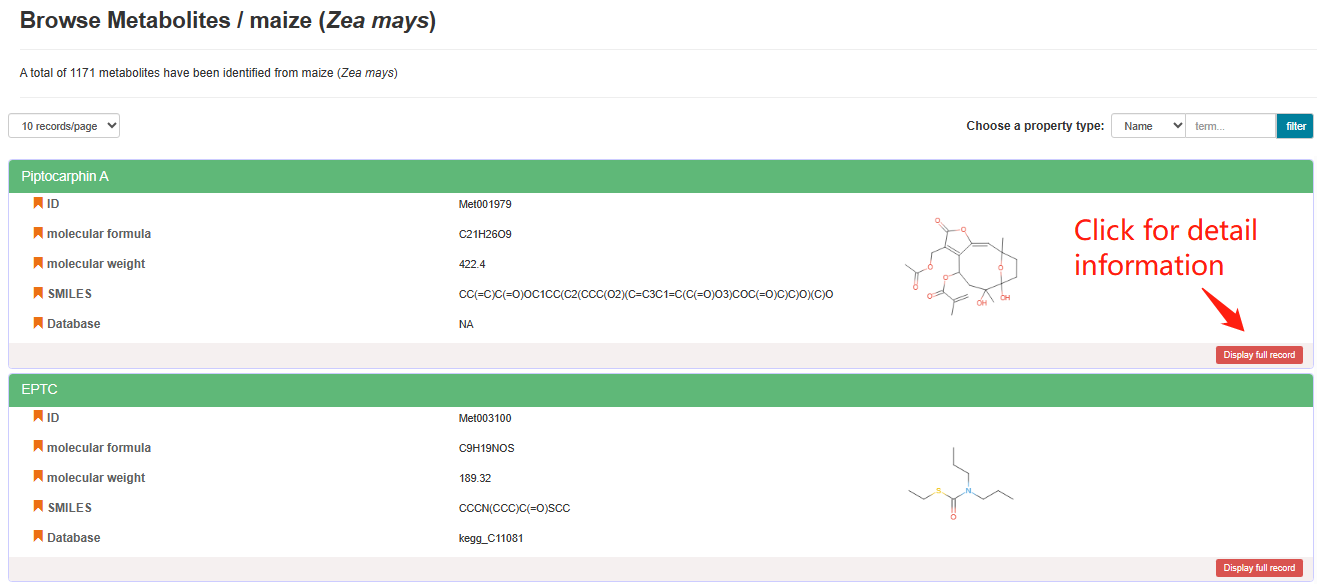
Browse Metabolites of a specific crop
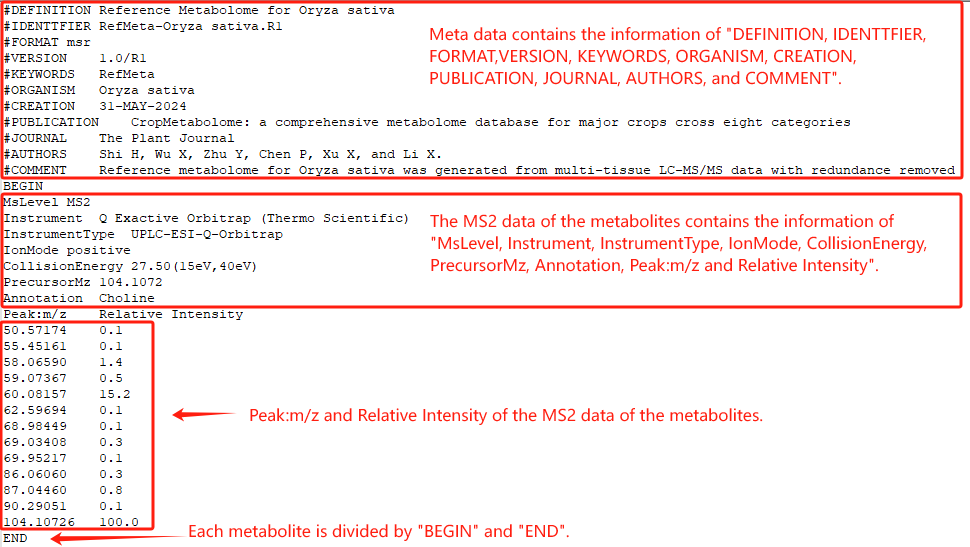
RefMeta-*.R1.msr file example
2 Browse metabolites for all crops
Usage
Users can click on the "Metabolite List" bar to access the Metabolite List webpage. The Metabolite List webpage displays the identified metabolites from all available crops in our database. Detailed information about a specific metabolite can be accessed by clicking the "Display full record" button.
Entrance for browsing crop metabolites
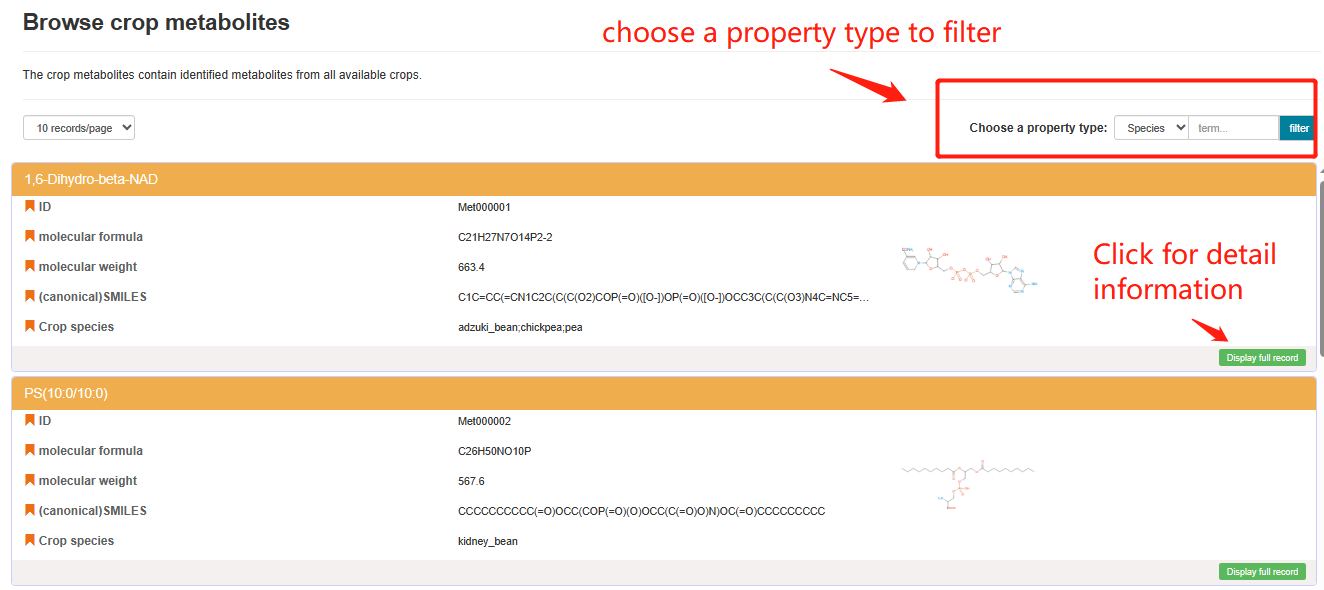
Browse crop metabolites
3 Browse pesticide
Usage
Users can click on the 'Pesticide List' bar to access the Pesticide List webpage. The Pesticide List webpage displays the pesticidal compounds collected in our database. Detailed information about a specific pesticide can be accessed by clicking the 'Display full record' button.
Entrance for browsing pesticides
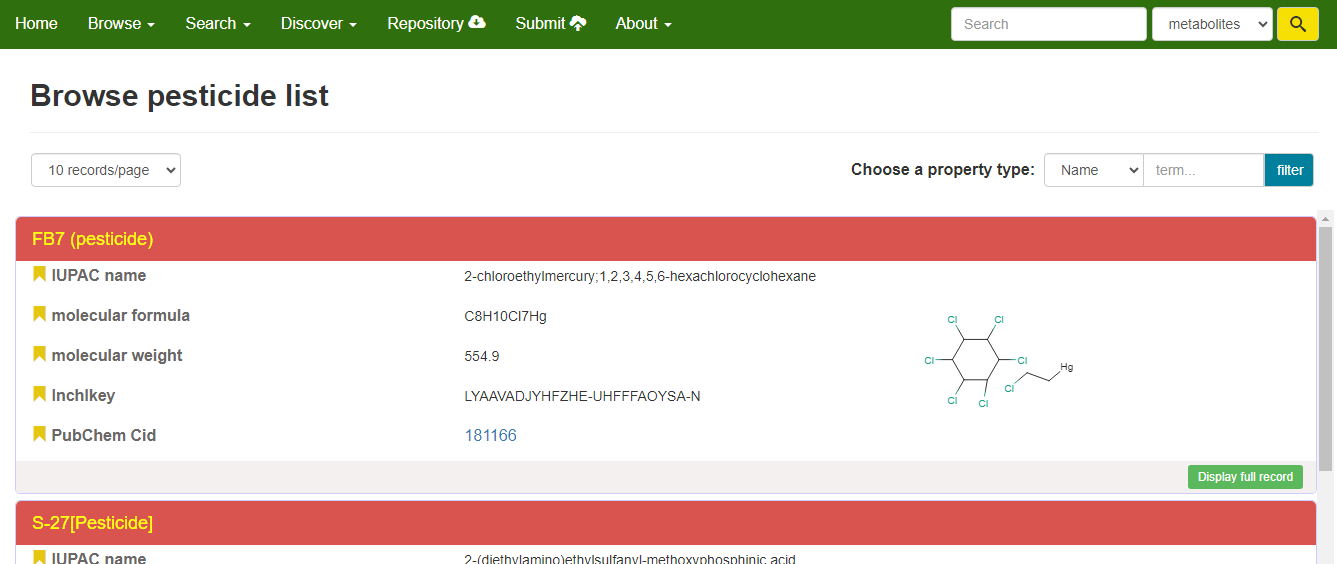
Browse pesticides
Search
In the searching metabolites module under “Search”, users are able to specify the domain you want to query. You can search by structure, compound weight range or advanced query to access metabolites and pesticides.
1 Structure Query
Usage
Users can draw a molecular structure on this page and then do an exact search to find records of related metabolites and pesticides.
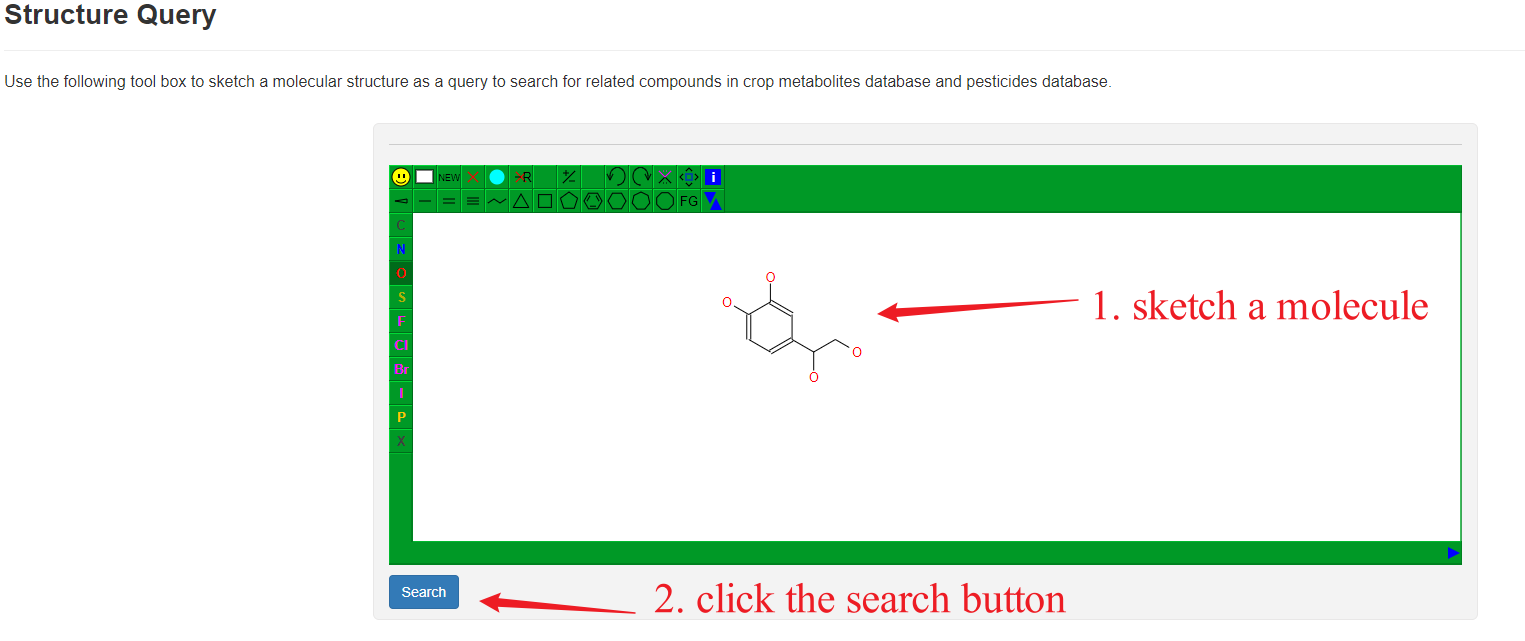
Overview of Structure Quary
2 Molecular Weight Query
Usage
The molecular weight query allows user to set the range of the molecule weight of interest to find records of related metabolites and pesticides.
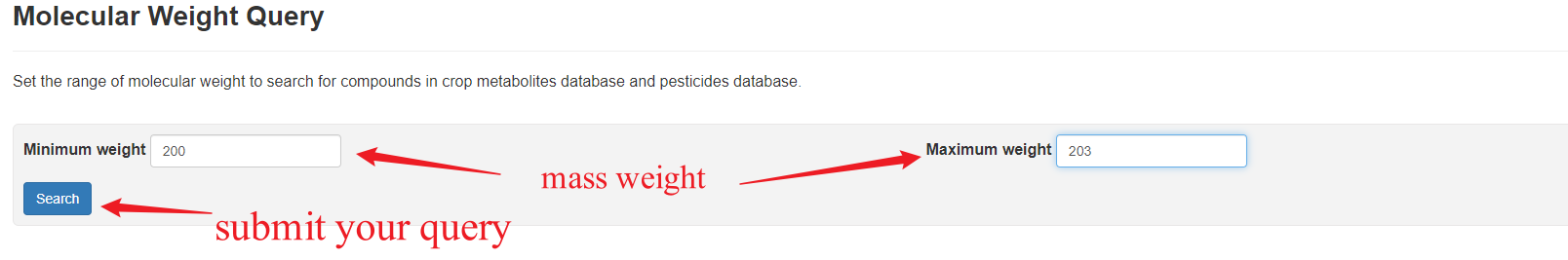
Overview of Molecular Weight Query
3 Advanced Query
Usage
Combined Query enables users to search for a metabolite or pesticide of their interest by specifying structural properties and molecular descriptors. Users can use one or combination of multiple of these options in "Name, Molecular Formula, ID, SMILES, InChI, Metabolite Class, Crop Species" to customize their search. After clicking the "Search" button, a new webpage will display matched metabolites and pesticides.
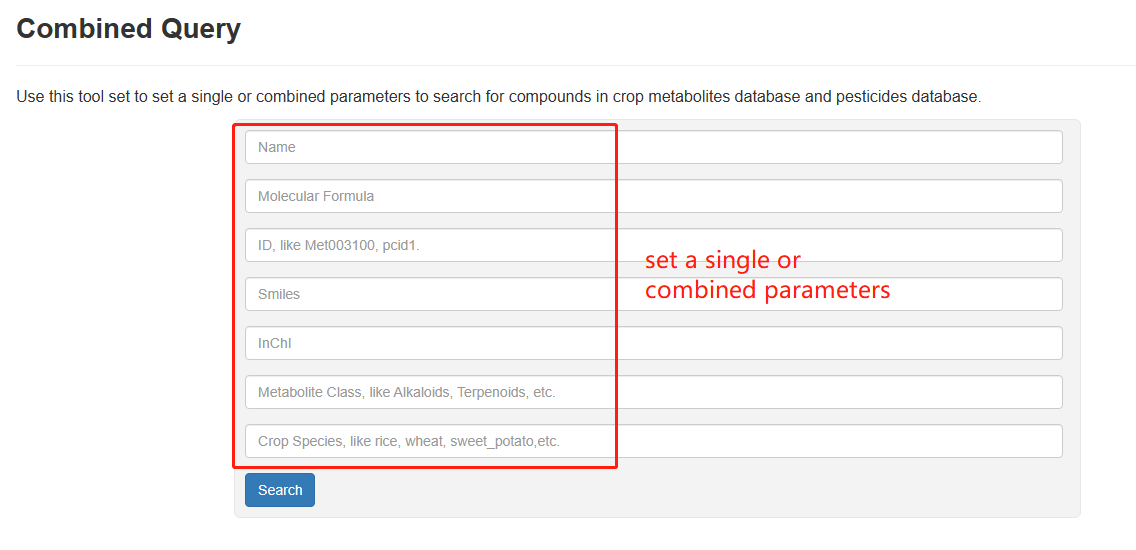
Overview of Combined Quary
4 LC-MS Query
Usage
LC-MS Query allows users to search against a MS2T library of chosen crop species uses one or multiple m/z values of precursor ions. Clicking the "Load Sample" button will automatically fill in a sample data. In this tool, one or multiple m/z values of precursor ions from sample MS spectra are manually entered in the text box. Then users can set the parameter of m/z tolerance, ion mode, and sample species, before query is executed by clicking the ‘search’ button.
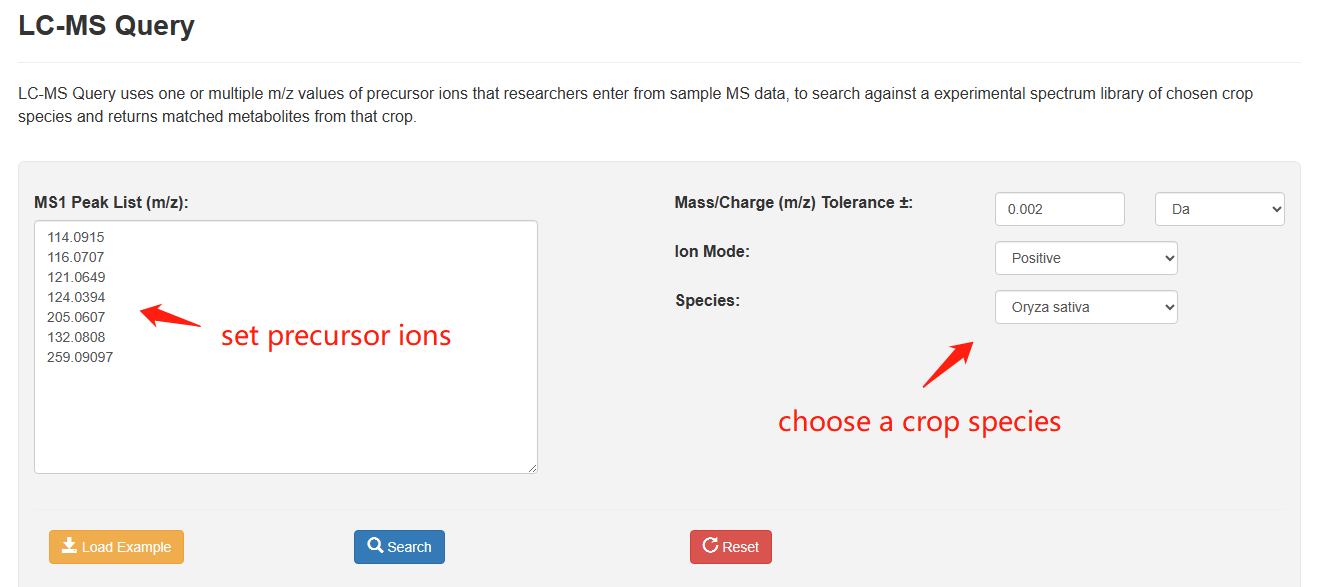
Overview of LC-MS Query
5 LC-MS/MS Query
Usage
LC-MS/MS Query allows users to use MS1 data (Parent Ion m/z) and MS2 data (Fragment Ions m/z and Intensity values) entered, to search against either the experimental spectral library (default), or both the experimental spectral library and the in silico spectral library (check select box). Clicking the "Load Sample" button will automatically fill in a sample data. Notably, in the returned results, the matched records are ordered with the similarity scores computed using the INCOS algorithm. It returns all matched reference compounds for the paired MS1/MS2 data of your interest.
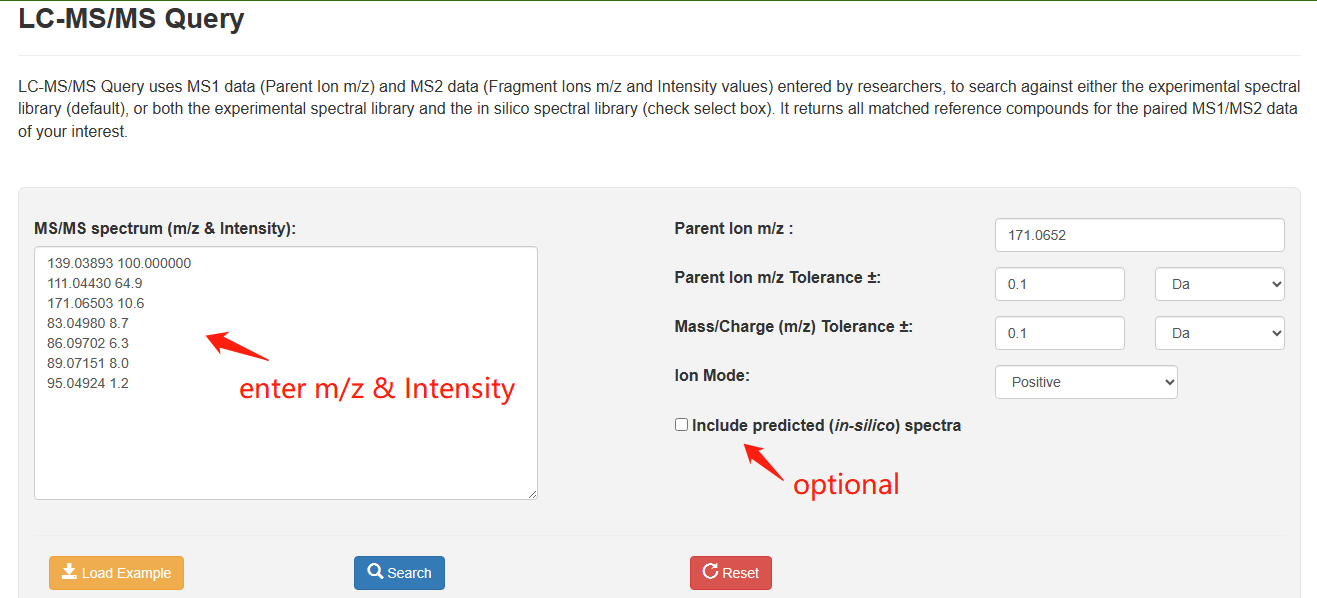
Overview of LC-MS/MS Query
Discover
The metabolite identification module under CropMetabolome "Discover" will be a good choice if you want to perform metabolite annotation or pesticide detection in crop samples.
1 CropRefMetaBlast
Usage
For a sample LC-MS datafiles in standard formats (mzML, mzXML, or mzData), CropRefMetaBlast function allows users apply our pipeline to perform metabolite annotation on the sample. You need to fill in the contact information and set the parameters for ion mode and the crop species accurately. Once the data is submitted and the analysis is complete, a web link to a result page will be returned. Users can use to retrieve the annotation results that include statistics of annotated peaks, categories of identified metabolites, and downloadable files for all extracted MS/MS spectra, and annotation of non-redundant peaks.
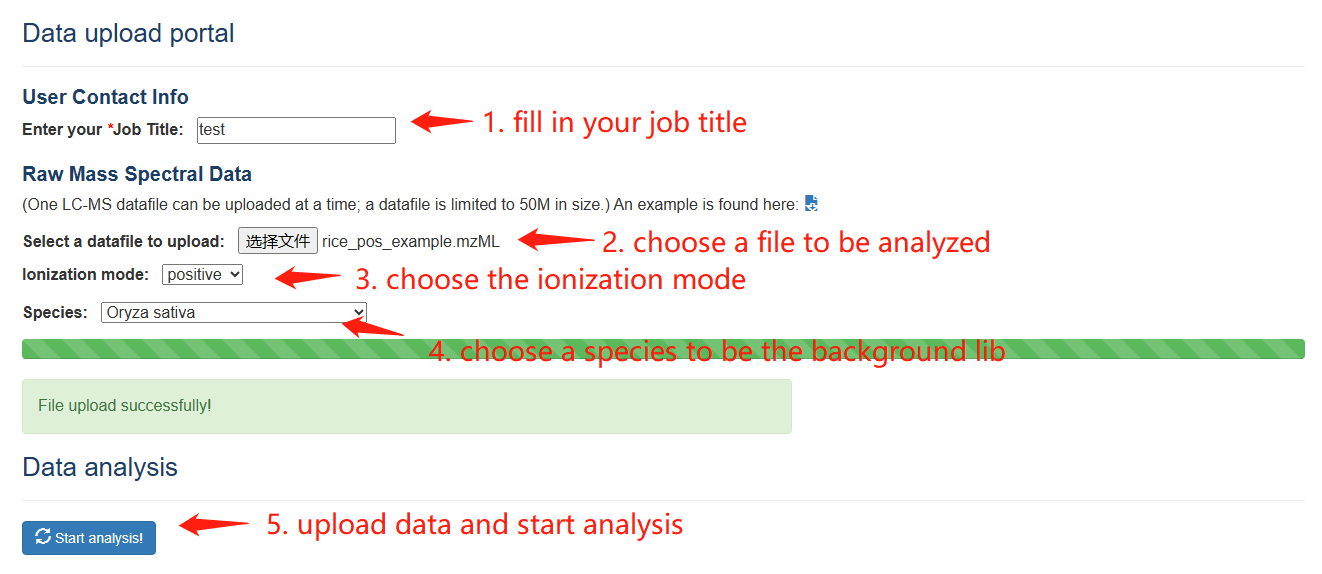
Overview of CropRefMetaBlast
2 CompoundLibBlast
Usage
For a sample LC-MS datafiles in standard formats (mzML, mzXML, or mzData), CompoundLibBlast function allows users apply our pipeline to perform metabolite annotation on the sample with our compound library. Like the CropRefMetaBlast page, users need to fill in the information of the Data upload portal carefully and a web link (valid for 72 hours) to a result page will be returned. Users can use to retrieve the annotation results that include statistics of annotated peaks, categories of identified metabolites, and downloadable files for all extracted MS/MS spectra, and annotation of non-redundant peaks.
Overview of CompoundLibBlast
3 PesticiDiscover
Usage
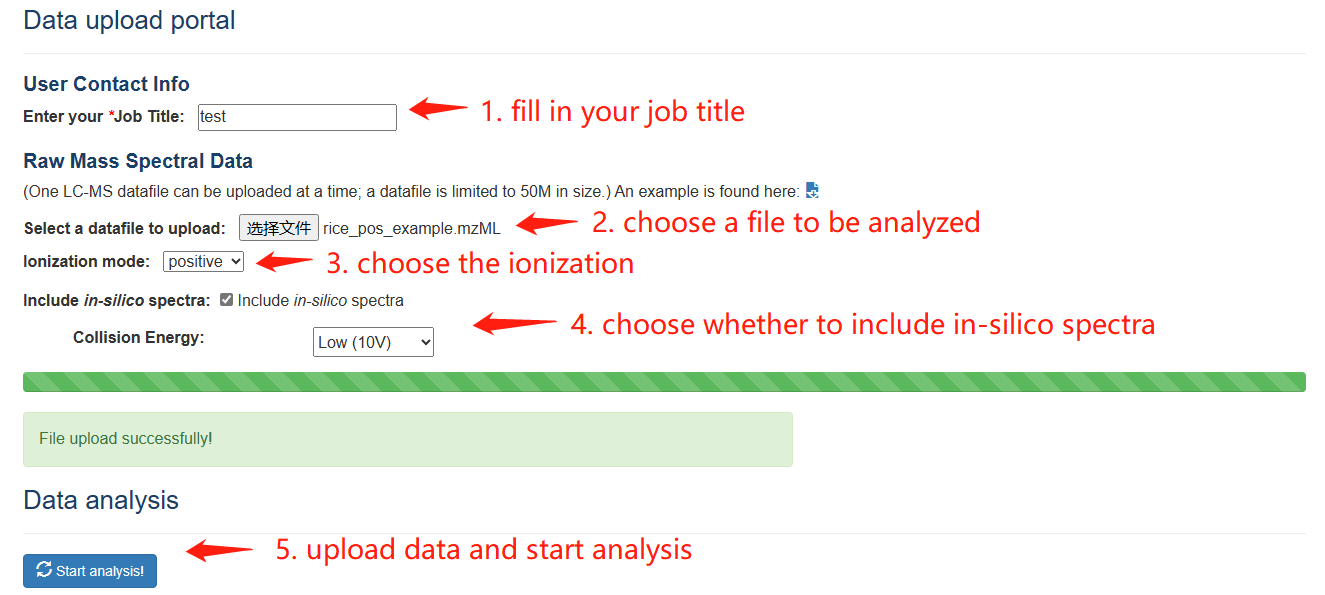
PesticiDiscover allows users to detect pesticide residues on the samples apply our pipeline. Like the CropRefMetaBlast page, users need to fill in the information of the Data upload portal carefully and a web link to a result page will be returned.
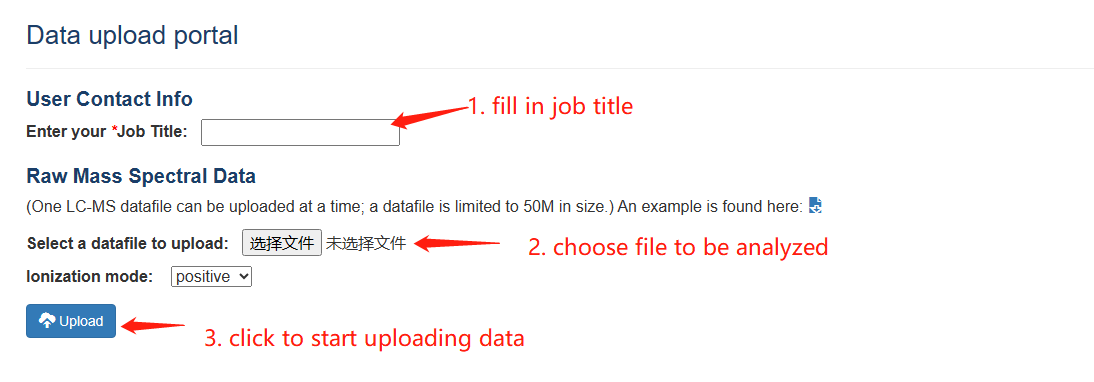
PesticiDiscover step 1-3
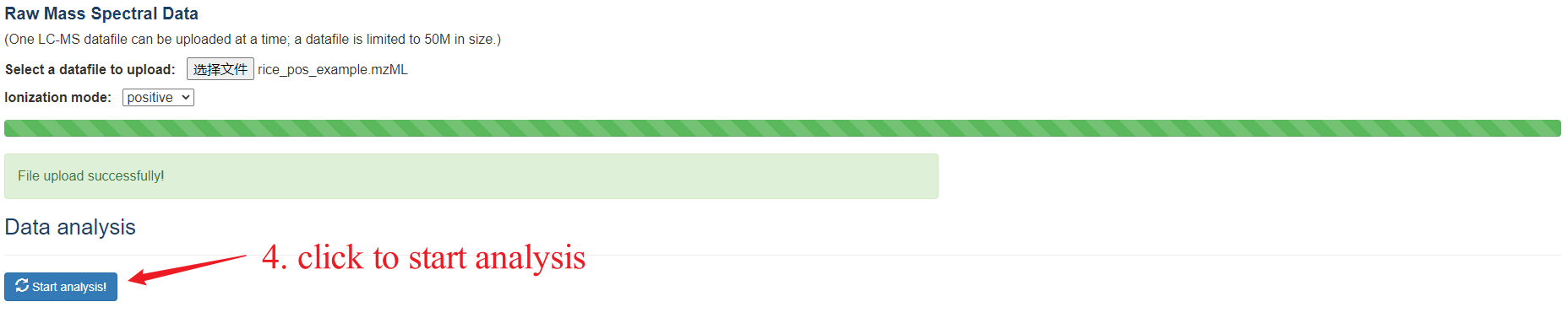
PesticiDiscover step 4
4 FlavoDiscover
Usage
FlavoDiscover allows users to use MS1 data (Precursor Ion m/z) and MS2 data (Fragment Ions m/z) that enter for the suspected precursor compounds, to search against the in-source fragment ions in the library of flavonoid motifs combined with neutral losses. Clicking the "Load Sample" button will automatically fill in a sample data. Results will be returned all matched flavonoids or flavonoid derivatives for the paired MS1/MS2 data after executing the query by clicking the "Search" button.
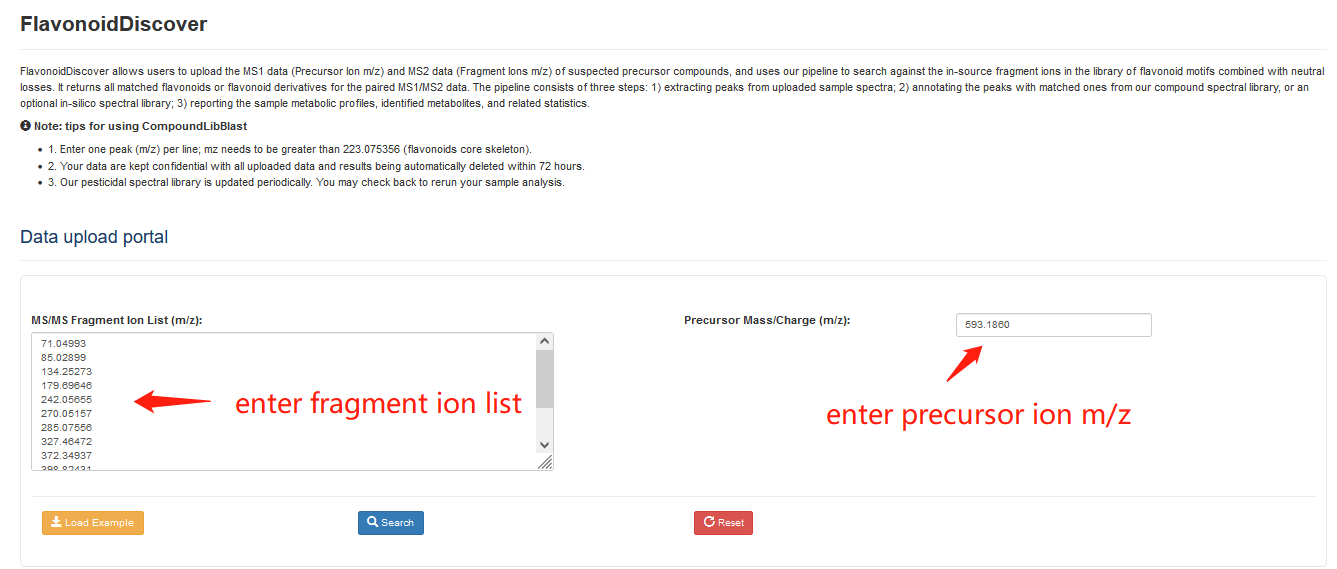
Overview of the FlavoDiscover
Repository
CropMetabolome is a public repository for sharing crop metabolomics data and users can freely and easily download raw data. In the webpage of “Repository” module, there is a download summary table that includes several data sections users can filter depends on their interest.
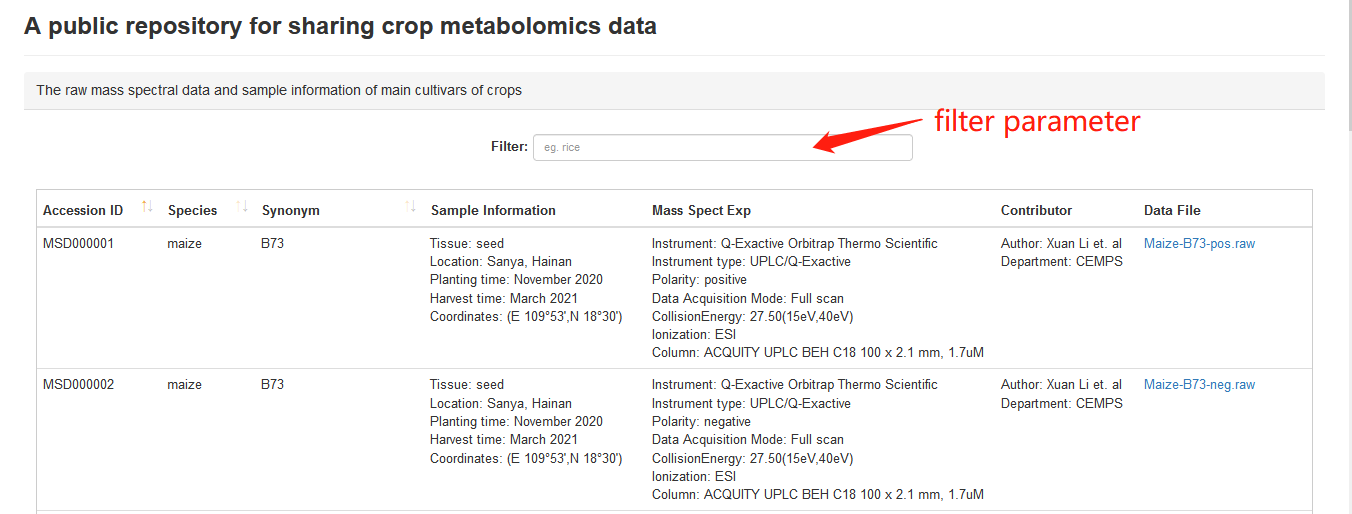
Overview of repository
Submit
In order to submit metabolomics data for CropMetabolome, you need to be first filled project-sample-metadata file and request a Project_ID by email. Your email will not be replied immediately, but after they have been confirmed by a reviewer. Once a Project_ID is assigned to you by email, you will be able to submit new data files, or update existing data files under the project you own. Your raw data will be found in “Repository” module within 15 days after you submit.
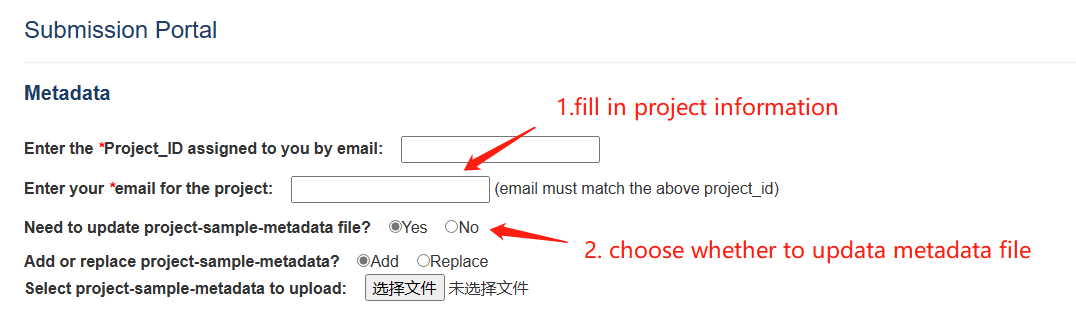
Submit step 1 & 2
Please do not use the "back" and "forward" button of your browser while doing a submit. After click on the "Submit " tab, you will see a progress bar for uploading data. If for some reasons your submit is interrupted, you can continue that in next time. You will see a green box on the new webpage where you can choose to continue to submit and this page will prompt the data files have been uploaded successfully.

Submit step 3
Details Page
1 Metabolites details
Metabolite Details
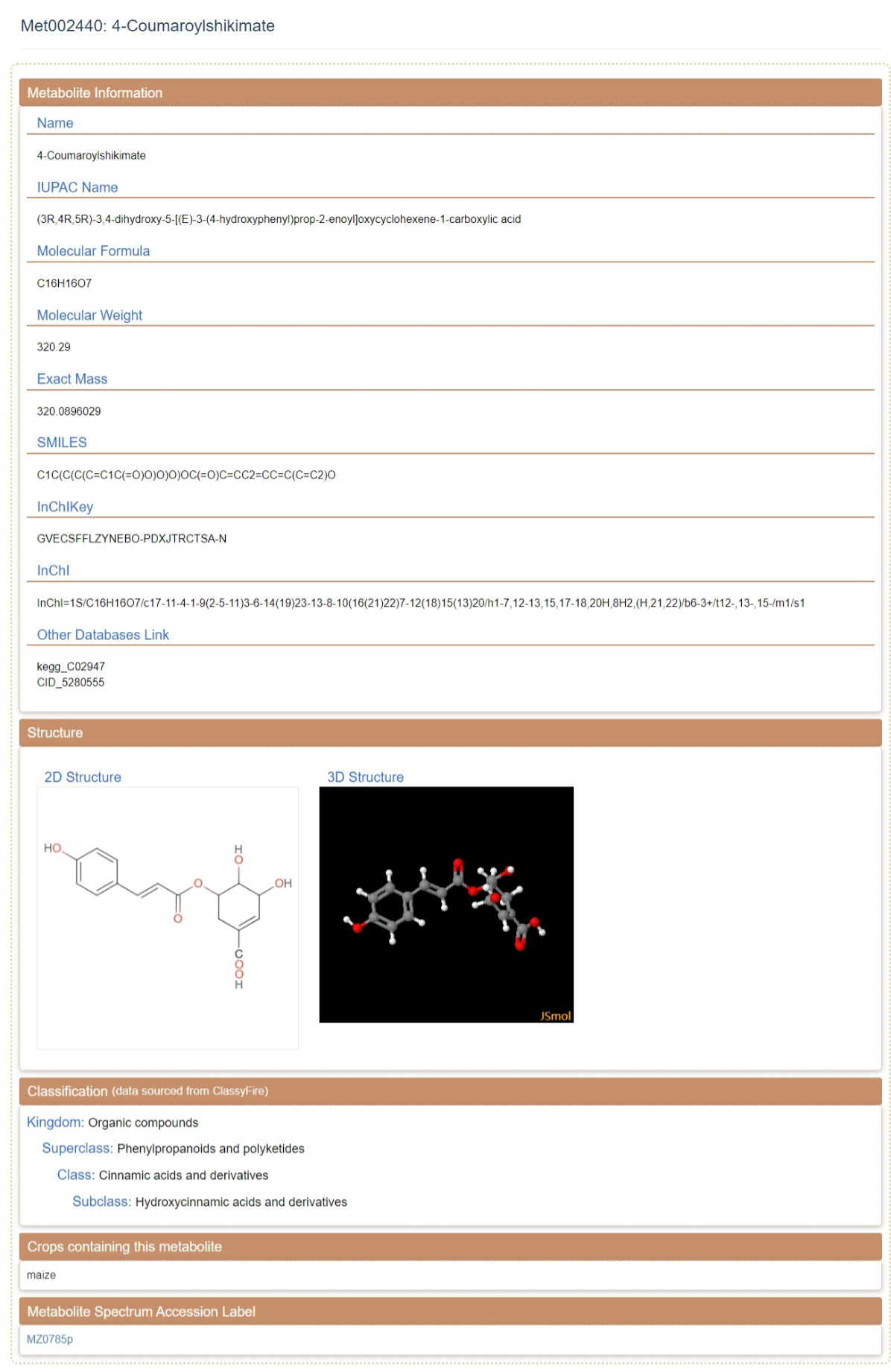
Metabolite Details
2 Spectrum details
Spectrum Details
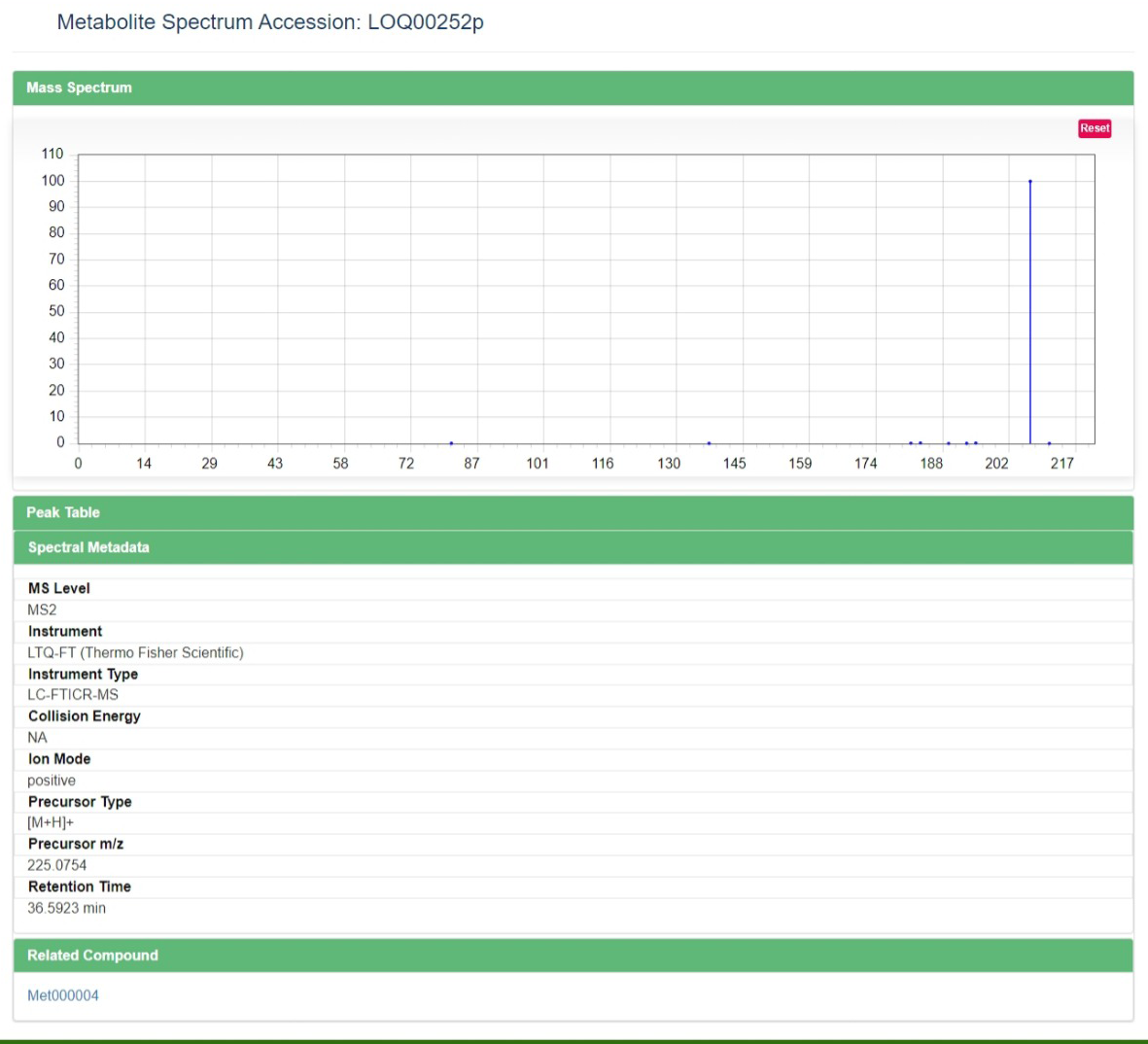
Spectrum Details
3 Pesticide details
Pesticide Details
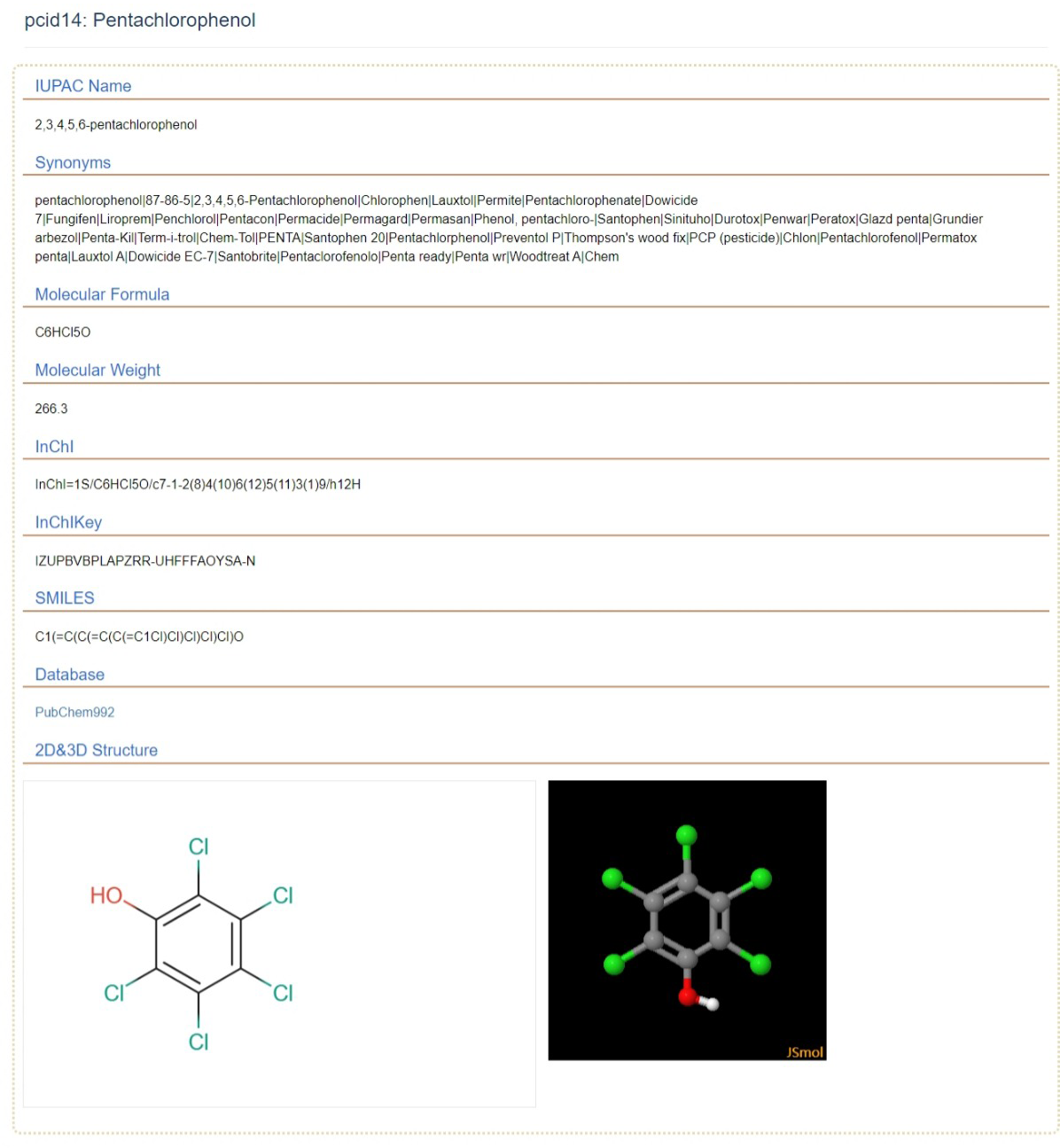
Pesticide Details
Terms and abbreviations
Terms and abbreviations commonly used in CropMetabolome.
LC-MS
liquid chromatography-mass spectrometry
LC-MS/MS
liquid chromatography tandem-mass spectrometry
MS2T library
MS/MS spectral tag (MS2T) library. MS2T libraries of CropMetabolome contains MS/MS spectra of known metabolites and unknown metabolites from crops.
MS2T Accession Label
Each MS2T accession was labeled in the format ‘RE0147p′ for example; this denotes the 147th spectrum (0147) derived from the library of rice (RE) extracts obtained in the positive ion mode (p, positive).
IUPAC
International Union of Pure and Applied Chemistry
InChI
IUPAC International Chemical Identifier
SMILES
Simplified Molecular Input Line Entry Specification
PubChem
a public database of chemicals and chemical information
CID
PubChem ID
KEGG
Kyoto Encyclopedia of Genes and Genomes